This is what it looks like when you do a genome assembly and run out of memory…
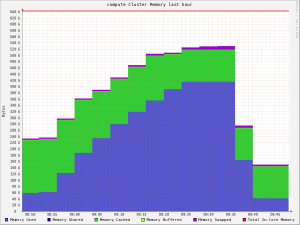
The machine in question actually has 384GB of RAM (not much, as far as machines which do genome assembly go!). Assembling a genome is like doing a massive puzzle; you need to have all the “pieces” in contiguous memory, and with the advances of genome sequencing technology there are always more “pieces” to put together!
The graph comes from Ganglia, a cluster monitoring tool we use on our high-performance computing cluster at ILRI, Nairobi. Ganglia won’t tell you when machines are down, slow, or whatever… but it’s great for a heads-up look at the health of your cluster (ie, why is there such a massive drop?), as well as hourly, daily, weekly trend graphs.
The culprit
As the graph was quite sharp to rise and fall, I assumed whatever job the user was running probably hadn’t ended amicably. A quick look at the machine’s dmesg verifies that assumption:
Out of memory: Kill process 6447 (velvetg) score 981 or sacrifice child
Killed process 6447, UID 901, (velvetg) total-vm:430619680kB, anon-rss:390267636kB, file-rss:4kBThe tool in question is velvet, a genome assembler. While genome assembly in general is memory intensive, I hear that velvet is especially so. 😉
Seems like kind of old software. Shouldn’t there be some GPU work happening for this type of stuff?
Nope, GPUs deal with highly-parallelizable problems, and genome assembly just isn’t there yet (or maybe General-Purpose GPU computing isn’t there yet). Genome assembly doesn’t even scale well on multiple cores because you incur high cost of synchronization between threads (“which pieces have you matched?” … “Ok, I’ve matched these ones.”).
A System with 400GB of RAM running out of memory! The first think that flashed in my mind was my 2GB laptop… 🙂 Can you limit Velvet’s memory usage?
Adam,
What Alan said about GPGPU compute is very true. Synchronization introduces major performance locks as threads communicate and stuff, and trust me, semaphores suck. Large kernels with locks for collision elision ( which re-introduces the problem you’re trying to avoid in the first place, a race condition). That stuff called Amdahl’s law kicks you in the jaw everytime you rework code to parallelism. And as is often the case in such software, the overheads (sync locks, atomics, etc) outweigh the performance boost you’d have gained IF (and IF) your code scales that well. Sure, the likes of OpenCL and CUDA are stepping in, but there’s only so much you can do for parallelism that would be deemed feasible enough to justify reworking all your code for parallelism only. Even OpenMPI isn’t exempted.
Conclusion: If your code is embarrassingly parallel ( and the code blocks don’t need to talk to each other), e.g graphics, fluid mechanics, ray scattering and physics rendering, go parallel.
However, in such cases where code blocks constantly exchange status updates and inter-process communication is mandatory (such as genome assembly), there be dragons when parallelism is called for.